
Volume 23, Number 9—September 2017
Dispatch
Norovirus in Bottled Water Associated with Gastroenteritis Outbreak, Spain, 2016
On This Page
Albert Blanco1, Susana Guix1, Noemí Fuster, Cristina Fuentes, Rosa Bartolomé, Thais Cornejo, Rosa Maria Pintó, and Albert Bosch
Abstract
In April 2016, an outbreak of gastrointestinal illness (4,136 cases) occurred in Catalonia, Spain. We detected high levels of norovirus genotypes I and II in office water coolers associated with the outbreak. Infectious viral titer estimates were 33–49 genome copies/L for genotype I and 327–660 genome copies/L for genotype II.
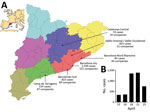
Figure. Waterborne norovirus outbreak in Catalonia, Spain, April 15–25, 2016 (n = 4,136 cases). A) Geographic distribution of the number of cases and affected companies in the Catalonian Health regions. Inset shows...
During April 11–25, 2016, a total of 4,136 cases of gastroenteritis were reported by the Public Health Agency of Catalonia (ASPCAT; Figure, panel A). A case-patient was defined as an exposed person who had vomiting or diarrhea (3 or more loose stools within 24 hours) and >2 of the following: nausea, abdominal pain, or fever (≥37.8°C). Six patients required hospitalization.
The epidemiologic investigation conducted by the ASPCAT pointed toward an association of the outbreak with drinking bottled spring water from office water coolers; the water had been bottled at a source in Andorra (M. Jané-Checa and A. Martínez-Mateo, Public Health Agency of Catalonia, pers. comm., 2016 Sep 1). Compared with other modes of transmission such as food or person to person, norovirus outbreaks associated with drinking water are rare in developed countries (1). On April 15, 2016, as a precautionary measure, the company producing the bottled water recalled >6,150 containers of water of suspected quality that had already been distributed to 925 companies. The water complied with all requirements of the European Commission directive on the exploitation and marketing of natural mineral waters (2), but these requirements do not include any virologic determination.
The Spanish Authority for Consumption, Food Safety, and Nutrition reported the outbreak at the national (http://www.aecosan.msssi.gob.es/AECOSAN/web/seguridad_alimentaria/ampliacion/gastroenteritis_agua_envasada.htm) and European (Rapid Alert System for Food and Feed, RASFF, expedient 2017/0469, https://webgate.ec.europa.eu/rasff-window/portal/?event = notificationDetail&NOTIF_REFERENCE = 2016.0469) levels. The numbers of cases reported by the ASPCAT peaked on April 18 and 21 (Figure, panel B), and the ASPCAT declared the outbreak over on April 25.
As part of the epidemiologic investigation of this outbreak, we took samples from four 19-L water coolers in 2 offices in the Barcelona metropolitan area, from which affected persons had drunk. We collected samples 1 and 2 on April 15 from 2 water coolers in 1 office, from which 36 cases had been reported. A private company provided samples 3 and 4, from 2 water coolers in a different office with an unknown number of cases, on April 20. We tested all samples immediately upon receipt at our laboratory. We used positively charged glass wool and polyethylene glycol precipitation for virus concentration. Sample volumes ranged from 2.0 L to 7.8 L; we reduced each sample to a final volume of 7 mL, as described previously (3). We extracted total RNA from 0.5 mL of the concentrates with the NucliSens miniMAG magnetic system (BioMérieux, Marcy-l’Étoile, France) and eluted the samples in 100 µL of elution buffer, following the manufacturer’s specifications. We performed a standardized 1-step real-time TaqMan reverse transcription PCR (RT-qPCR; Ultrasense, Invitrogen Life Technologies, Barcelona, Spain), in which we used 5 µL of extracted RNA to determine the number of genome copies per liter of human norovirus genogroup I (GI) and genogroup II (GII) (4–7). We monitored virus/nucleic acid extraction and enzyme efficiencies as previously described; we used double-stranded DNA plasmids containing the target sequences as standards (8).
We detected high RNA levels for norovirus GI and GII, around 103 and 104 genome copies/L, in 2 of the 4 water cooler samples concentrated by glass wool filtration and polyethylene glycol precipitation (Table). Because molecular methods are unable to discern between infectious and noninfectious particles, we predicted the infectivity of norovirus in the concentrated samples by treating the samples with the nucleic acid intercalating dye PMA propidium monoazide; (50 µmol/L) and Triton X surfactant (0.5%) before RT-qPCR; this enabled us to distinguish between virions with intact and altered capsids (9). Following this approach, estimated infectious levels in the 2 positive samples were 49 and 327 genome copies/L for norovirus GI and 33 and 660 genome copies/L for norovirus GII (Table).
Given the large number of persons involved in the outbreak and the reported 50% human infectious dose for norovirus of 18–1,300 particles (10,11), the high genome copy values in the water samples were not unexpected. In addition, the proportion of intact (infectious) virions in the water samples, estimated through PMA/Triton treatment before RT-qPCR assays, represented 0.3%–5.6% of the total number of physical particles, which was enough to cause gastrointestinal illness (10,11).
We assayed the presence of enteroviruses, astroviruses, sapoviruses, rotaviruses, adenoviruses, and hepatitis A virus in the 4 water samples by using commercial RT-qPCR kits (Viasure, Certest Biotec SL, Zaragoza, Spain), with negative results. We attempted genotyping of noroviruses in samples 1 and 2 using a semi-nested RT-PCR protocol with specific primers for GI and GII. We performed the first PCR with primers COG1F and G1SKR for GI and COG2F and G2SKR for GII (6,12). For the second PCR, we used primers G1SKF and G1SKR for GI and G2SKF and G2SKR for GII (12). We assigned genotypes based on clustering with reference strains from the sequence database of the European network NoroNet and norovirus genotyping tool (13). We detected a single sequence corresponding to genotype GII.4/Sydney/2012 (GenBank accession no. KX816644) in samples 1 and 2. Additionally, MiSeq next-generation sequence analysis (Illumina, San Diego, CA) of the amplified product confirmed the sole presence of genotype GII.4/Sydney/2012 (data not shown).
Although some fecal samples from persons who worked at the office from which water samples 1 and 2 were obtained contained genotypes GI.2 (n = 10) and GII.17 (n = 11) but not GII.4/Sydney/2012, we isolated genotypes GII.4/Sydney/2012 (n = 1), GI.2 (n = 1), GII.17 (n = 1), and GII.2 (n = 1) from fecal samples from persons from a different office who exhibited the same gastrointestinal symptoms after drinking water supplied by the same company (data not shown). We hypothesize that the spring water was contaminated by all 4 strains (GI.2, GII.2, GII.4, and GII.17) but levels of viral contamination for each genotype were not homogeneous in all bottled coolers. We may have detected only the GII.4 genotype in water samples 1 and 2 because of a higher concentration of this specific genotype or because of bias caused by the sampling, concentration, and molecular detection procedures. Finally, several reasons could explain why we did not find any GII.4/Sydney/2012 or GII.2 genotypes among the fecal samples from persons from the office that provided samples 1 and 2, including the existence of immune status among the exposed persons or differences in the proportion of infectious/physical particles between the different types.
We describe quantitative detection of norovirus in bottled water. Previously, several brands of mineral water were reported to contain norovirus, but the findings were later attributed to laboratory contamination with control reagents (14,15). One limitation of our study is the low number of water samples analyzed. Four days after the onset of the outbreak, the company recalled all batches of water and water coolers of suspected quality, which hampered the collection of a larger number of samples for analysis.
The cause of the water contamination remains to be elucidated. However, the high number of affected persons from 381 offices that received water coolers, and the many different genotypes found in some patients’ fecal specimens, point toward sewage pollution of the spring aquifer. Aquifer pollution was acknowledged by the Andorra Ministry of Health and Welfare, and further use of the spring was banned.
This large outbreak suggests that the management of microbial risks of commercially produced mineral waters, universally based solely on bacterial parameters, could benefit from additional analysis for relevant viral pathogens such as norovirus. However, the substantial costs incurred in developing, enhancing, and managing virus surveillance systems call for a balanced approach to keep both the cost and the time required for the analyses within feasibility limits.
Mr. Blanco is a PhD student at the Department of Genetics, Microbiology, and Statistics of the University of Barcelona in Barcelona, Spain. His research interests include molecular diagnostics and epidemiology of enteric viruses in clinical and environmental samples. Dr. Guix is an associate professor in the Department of Genetics, Microbiology and Statistics of the University of Barcelona, and is a member of the scientific committee of the Spanish Authority for Consumption, Food Safety, and Nutrition. Her research interests include epidemiology and food safety studies on viruses causing gastrointestinal diseases and hepatitis.
Acknowledgments
We are grateful to M. Jané-Checa and A. Martínez-Mateo for providing epidemiological data and useful discussion and to J. Quer and J. Gregori for next-generation sequencing and bioinformatic assistance. We acknowledge the useful collaboration with A. Canals.
This work was supported in part by projects Food-FP7-311846 (European Union) and XRB-Biotechnology Reference Network (Generalitat de Catalunya). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Bitler EJ, Matthews JE, Dickey BW, Eisenberg JN, Leon JS. Norovirus outbreaks: a systematic review of commonly implicated transmission routes and vehicles. Epidemiol Infect. 2013;141:1563–71. DOIPubMed
- European Union. Directive 2009/54/EC of the European Parliament and of the Council of 18 June 2009 on the exploitation and marketing of natural mineral waters. Recast. 2662009: L 164. Luxembourg; 2009.
- Pérez-Sautu U, Sano D, Guix S, Kasimir G, Pintó RM, Bosch A. Human norovirus occurrence and diversity in the Llobregat river catchment, Spain.Environ Microbiol. 2012;14:494–502. DOIPubMed
- da Silva AK, Le Saux JC, Parnaudeau S, Pommepuy M, Elimelech M, Le Guyader FS. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl Environ Microbiol. 2007;73:7891–7. DOIPubMed
- Svraka S, Duizer E, Vennema H, de Bruin E, van der Veer B, Dorresteijn B, et al. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J Clin Microbiol. 2007;45:1389–94. DOIPubMed
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–57. DOIPubMed
- Loisy F, Atmar RL, Guillon P, Le Cann P, Pommepuy M, Le Guyader FS. Real-time RT-PCR for norovirus screening in shellfish. J Virol Methods. 2005;123:1–7. DOIPubMed
- International Organization for Standardization (ISO). ISO/TS 15216–1:2013. ISO/TS 15216. Microbiology of food and animal feed–Horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR. Geneva: The Organization; 2013.
- Fuster N, Pintó RM, Fuentes C, Beguiristain N, Bosch A, Guix S. Propidium monoazide RTqPCR assays for the assessment of hepatitis A inactivation and for a better estimation of the health risk of contaminated waters. Water Res. 2016;101:226–32. DOIPubMed
- Teunis PFM, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, et al. Norwalk virus: how infectious is it? J Med Virol. 2008;80:1468–76. DOIPubMed
- Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, et al. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis. 2014;209:1016–22. DOIPubMed
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, et al. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–14. DOIPubMed
- Kroneman A, Vennema H, Deforche K, v d Avoort H, Peñaranda S, Oberste MS, et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol. 2011;51:121–5. DOIPubMed
- Beuret C, Kohler D, Lüthi T. Norwalk-like virus sequences detected by reverse transcription-polymerase chain reaction in mineral waters imported into or bottled in Switzerland. J Food Prot. 2000;63:1576–82. DOIPubMed
- Sanchez G, Joosten H, Meyer R, Beuret C. Presence of norovirus sequences in bottled waters is questionable. Appl Environ Microbiol. 2005;71:2203–5, author reply 2203–5. DOIPubMed
Figure
Table
Cite This Article1These authors contributed equally to this article.






















.png)











No hay comentarios:
Publicar un comentario